- Department of Neurosurgery, Barrow Neurological Institute, St. Joseph's Hospital and Medical Center, Phoenix, Arizona, USA
- Department of Neurosurgery, Irkutsk State Medical University, Irkutsk, Russia
- College of Medicine-Phoenix, University of Arizona, Phoenix, Arizona, USA
Correspondence Address:
Mark C. Preul
Department of Neurosurgery, Barrow Neurological Institute, St. Joseph's Hospital and Medical Center, Phoenix, Arizona, USA
DOI:10.4103/sni.sni_55_18
Copyright: © 2018 Surgical Neurology International This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.How to cite this article: Arpan Patel, Evgenii Belykh, Eric J. Miller, Laeth L. George, Nikolay L. Martirosyan, Vadim A. Byvaltsev, Mark C. Preul. MinION rapid sequencing: Review of potential applications in neurosurgery. 10-Aug-2018;9:157
How to cite this URL: Arpan Patel, Evgenii Belykh, Eric J. Miller, Laeth L. George, Nikolay L. Martirosyan, Vadim A. Byvaltsev, Mark C. Preul. MinION rapid sequencing: Review of potential applications in neurosurgery. 10-Aug-2018;9:157. Available from: http://surgicalneurologyint.com/surgicalint-articles/minion-rapid-sequencing-review-of-potential-applications-in-neurosurgery/
Abstract
Background:Gene sequencing has played an integral role in the advancement and understanding of disease pathology and treatment. Although historically expensive and time consuming, new sequencing technologies improve our capability to obtain the genetic information in an accurate and timely manner. Within neurosurgery, gene sequencing is routinely used in the diagnosis and treatment of neurosurgical diseases, primarily for brain tumors. This paper reviews nanopore sequencing, an innovation utilized by MinION and outlines its potential use for neurosurgery.
Methods:A literature search was conducted for publications containing the keywords of Oxford MinION, nanopore sequencing, brain tumor, glioma, whole genome sequencing (WGS), epigenomics, molecular neuropathology, and next-generation sequencing (NGS). In total, 64 articles were selected and used for this review.
Results:The Oxford MinION nanopore sequencing technology has had successful applications within clinical microbiology, human genome sequencing, and cancer genotyping across multiple specialties. Technical details, methodology, and current use of MinION sequencing are discussed through the prism of potential applications to solve neurosurgery-related scientific and diagnostic questions. The MinION device has proven to provide rapid and accurate reads with longer read lengths when compared with NGS. For applications within neurosurgery, the MinION device is capable of providing critical diagnostic information for central nervous system (CNS) tumors within a single day.
Conclusions:MinION provides rapid and accurate gene sequencing with better affordability and convenience compared with current NGS methods. Widespread success of the MinION nanopore sequencing technology in providing accurate, rapid, and convenient gene sequencing suggests a promising future within research laboratories and to improve care for neurosurgical patients.
Keywords: DNA, MinION, nanopore, neurosurgery, sequencing, tumor
INTRODUCTION
Since its introduction, gene sequencing has revolutionized the understanding of human genetics and the practice of medicine. The advent of sequencing made way for landmark achievements such as the Human Genome Project.[
Gene sequencing has become critical to multiple aspects of neurosurgery including the diagnosis, treatment, and evaluation of central nervous system (CNS) tumors.[
MATERIALS AND METHODS
This study does not endorse any specific corporate technology and has no marketing or financial relationship with any corporate entity or trademarked technology named in this paper. This study received no outside funding. An electronic literature search was conducted using the National Library of Medicine for publications containing the keywords of Oxford MinION, nanopore sequencing, brain tumor, glioma, whole genome sequencing (WGS), epigenomics, molecular neuropathology, third generation sequencing (TGS), and NGS. Bibliographies of select publications were additionally reviewed to complete the literature search. The articles were screened and selected based on the inclusion criteria for topics that examined the use of nanopore sequencing, particularly the Oxford MinION. Additional articles pertaining to the understanding of DNA sequencing were also included. In total, 64 articles were selected for review.
Gene sequencing
The first method of sequencing, known as Sanger Sequencing, was introduced in 1977 and remained the gold standard of sequencing until the popularization of second-generation sequencing (SGS), also named NGS, in 2005.[
Two major technologies exist under the umbrella of TGS: PacBio's single molecular real time (SMRT) sequencing and Oxford's Nanopore Technology (ONT). The latter technology, also called nanopore sequencing, was a concept first suggested in 1995 in a concerted effort by multiple research groups, notably including George M. Church.[
Oxford MinION device
Sequencing
Understanding nanopore technology and the molecular science behind the Oxford MinION may present a daunting task for clinicians removed from basic science research. In this paper, we present a simplified version. The MinION is a handheld 90 g device that can plug into any computer with a standard USB 3.0 port. The MinION functions by passing long sequences of DNA (8-20 kbp) through a pore within a small protein, “the nanopore,” that is embedded within a membrane. The single stranded DNA molecule (ssDNA) that passes through the nanopore can optionally contain a hairpin adaptor that physically connects the “template” strand and the “complimentary” strand [
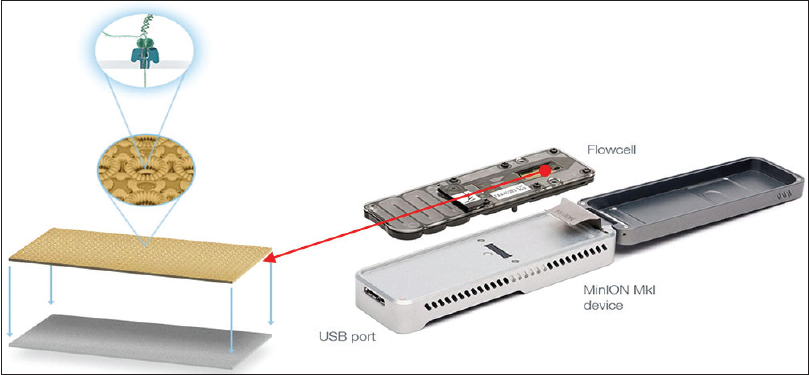
Figure 1
Miniature nanopore sequencing device with microscopic view of flowcell containing embedded nanopore[
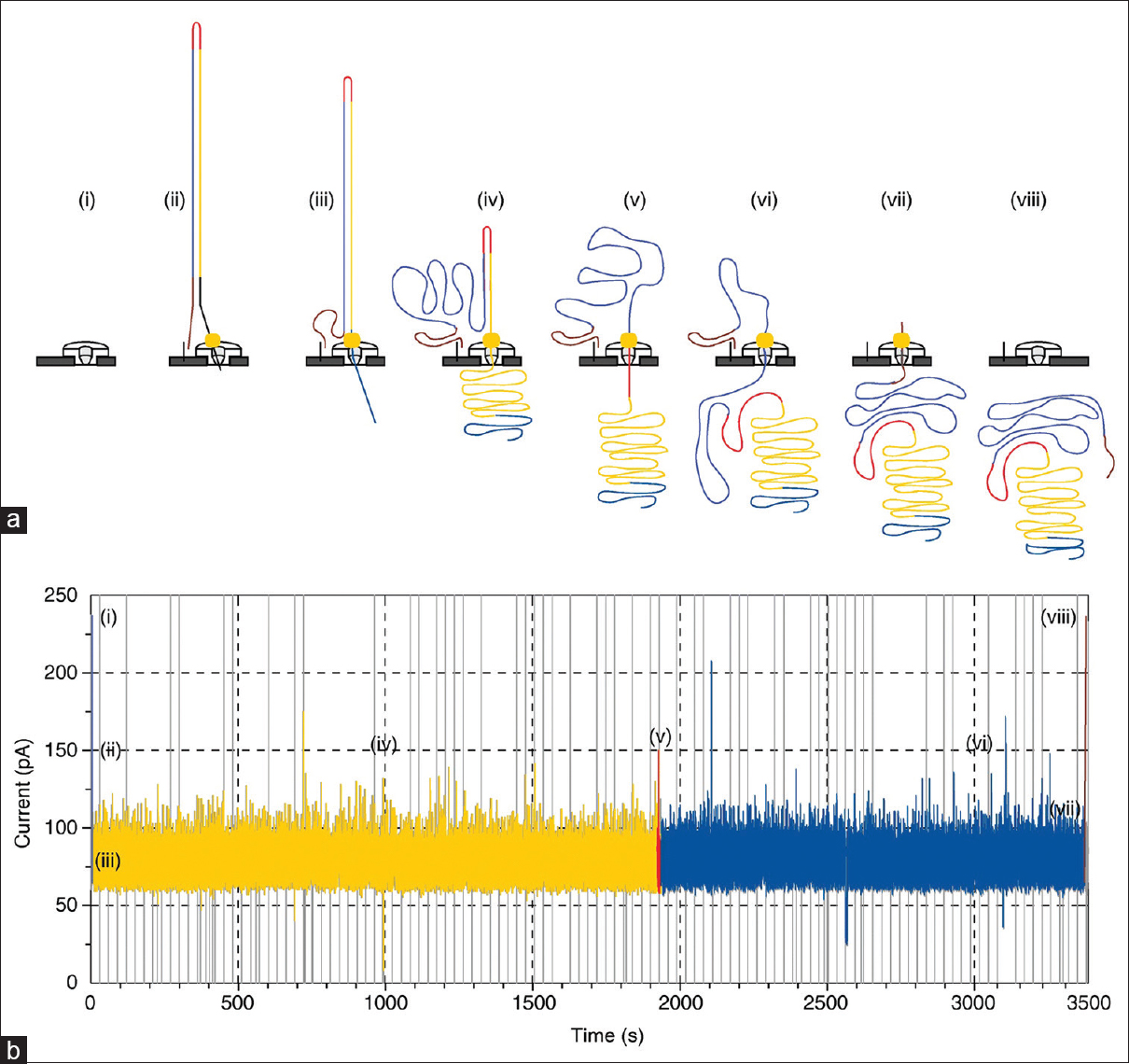
Figure 2
Principle of nanopore sequencing. (a) Visualization of 2 ssDNA strands connected by adaptor passing through the nanopore. (b) Graphical representation of raw data produced by MinION, also known as “squiggle plot”[
Basecalling
Once the device is plugged into a computer, it is operated with the MinKNOW software program. This program is responsible for acquiring and analyzing data, and providing the interface for additional device control. After the raw data, saved as a FAST5 file, are collected, it must undergo an analysis to be converted into a nucleotide sequence; this process is termed basecalling. There are multiple software platforms that are publicly available to complete the basecalling process, namely, Metrichor,[
Further analytics
Once the initial data are collected, further analysis can provide additional information regarding the presence of mutations, characterization of base repeats, and specific methylation patterns. Many of these specific analytical tasks require supplemental software to accomplish. To detect mutations within the targeted sequences, the template reads must be compared with a reference human genome. This can be accomplished using Galaxy,[
The detection of copy number variation is a complicated process that is prone to error. To maximize accuracy, multiple steps are typically involved in analyzing the DNA sequence. The sequence is first “normalized” using specific software such as QDNAseq[
Direct detection of methylation patterns of DNA is one of the unique strengths of nanopore sequencing. Specific studies have shown that the 5-methylcytosine (5-mC) modified base can be directly detected based on unique electrical signals detected by the MinION device without the need for additional chemical treatment. HMM models can be trained to detect specific methylation patterns and distinguish them from other base pairs. However, this technology is still under development and has shortcomings. Not all methylated bases can be detected, and when both methylated and unmethylated bases are present within the same k-mer, the detection system is inaccurate. Improvement in basecalling software can be expected to resolve these obstacles.[
Sample preparation
A DNA-containing sample needs to be treated with a variety of steps before it is ready to be analyzed with the MinION device. This process, named library preparation, has recently been simplified and is available in predesigned kits. The components and protocol included in each kit are designed to achieve a particular purpose desired by the investigator. Kits available for purchase online include Ligation Sequencing Kit 1D (SQK-LSK108), Rapid Barcoding Kit (SQK-RBK004), Rapid Sequencing Kit (SQK-RAD004), and 1D2 Sequencing Kit (SQK-LSK308). Kits that focus on preparation of a cDNA or RNA sample are also available. ONT continuously updates these kits, improving on the protocol and components to provide the best outcome and experience in library preparation. Intersection of computer programming and the electrical manipulation of fluids resulted in the creation of VolTRAX, a handheld automated library preparation device with consumable cartridges. The device allows the user to load samples into the cartridge and run a preprogrammed code that manipulates, moves, and mixes the liquid samples as desired. VolTRAX is designed to provide reproducible, portable, and simplified library preparation.[
Improvements in technology and throughput
The final accuracy of the nucleotide sequence is largely influenced by both the basecalling process and the internal chemistry of the nanopore. Basecalling software includes technology that uses deep and machine learning models to convert the raw electrical signal into a nucleotide sequence. Basecalling accuracy has rapidly improved between 2014 and 2017 with new software releases by ONT, such as Albacore v2.0.1, and third-party researchers publishing open-source software. Both Chiron and Albacore v2.0.1 are recently released programs (September 2017) that bypass error-prone steps to provide an improved accuracy in basecalling.[
Another aspect of the MinION pipeline that can significantly affect the throughput and accuracy of the sequencing is the flow cell. The flow cell is the consumable part of the MinION that contains the nanopore proteins and the Application-Specific Integrated Circuit (ASIC) sensor. The flow cell is also the site where the prepared DNA sample is directly added. Currently, there are two flow cells manufactured by ONT, the R9.4 and R9.5. Previous flow cells that have since been improved include the R7.3 and R9. The R9.4 flow cell is graded for all 1D experiments, while the R9.5 is compatible with both 1D and 1D2.[
Current applications
After only 4 years since its release, ONT's MinION has been utilized in a wide variety of applications, including WGS,[
Tumors
Recent (2016) World Health Organization (WHO) classifications for CNS tumors, for the first time, require molecular and genetic analysis for definitive diagnosis.[
The MinION device has also been used in other cancers, including leukemia,[
Microbiology
In addition to its application for gene profiling in cancer, the MinION has had considerable success in the identification of microbiology, WGS, SNP identification, and forensics. Multiple studies have utilized the MinION to identify and characterize specific pathogen species within known microbiological communities and clinical samples.[
Human genome sequencing
Current commercial and hospital laboratory-based methods of WGS are limited to NGS techniques. As research continues to implicate genetic mutations, specifically SNPs, in context of countless diseases, the need for genetic screening and WGS steadily rises. Advancement in precision medicine and personalized treatment plans are facilitated by research demonstrating actionable results and progress in the speed, cost, and accuracy of WGS. Within neurosurgery, many disease processes have been found to have a major genetic component. Recent reports on intervertebral disc disease suggest that up to 75% of the underlying etiology is attributed to genes.[
Nanopore sequencing has demonstrated the capability to accurately and rapidly sequence the whole genome of bacteria, eukaryotes, and viruses reliably.[
Jain et al. reported the use of the MinION in assembling a high consensus human genome.[
DISCUSSION
MinION in the context of neurosurgery
Based on the current applications of the MinION, its potential use in neurosurgery is diverse. Particularly for CNS tumors, integration of the MinION into the diagnostic pipeline promises to reduce costs and turn-around time for actionable results. The 2016 WHO classifications of CNS tumors require molecular profiling for final diagnosis. Common genes that delineate this classification include IDH, 1p/19q, SHH, WNT, TP53, and RELA.[
NGS panels directed at detecting critical mutations in CNS tumor tissue are routinely used in the hospital setting.[
Limitations
An identifiable challenge that will hinder the rapid and widespread use of portable nanopore sequencers is its competition with genomic services that provide “all-inclusive” (sample preparation, sequencing, computational analysis, and interpretation) sequencing in a timely manner, with medical center samples submitted by mail. These companies, providing WGS services, offer a price per sample that is roughly comparable with the entire cost of a portable sequencer.[
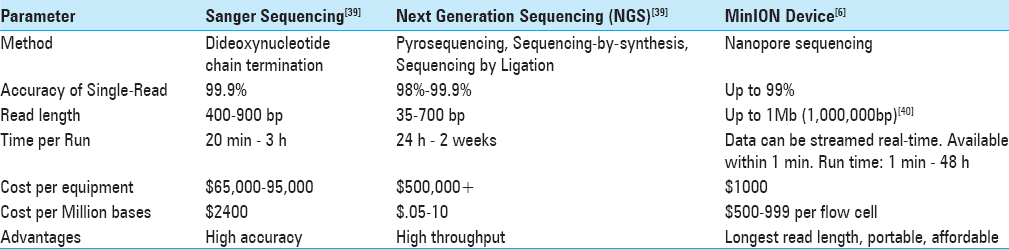
Comparison of the NGS MinION with the previous generation and more common sequencing methods is summarized in
CONCLUSION
Low cost, portability, speed, and versatility make MinION a promising tool for low-resolution, rapid nucleic acid sequencing. As the accuracy of MinION increases with improved chemistry and basecalling software, we predict that nanopore sequencing may become a widely used tool for rapid gene profiling. Based on the first reports available so far, nanopore sequencing technology has a potential to improve the molecular diagnostic pipeline for CNS tumors, infectious diseases, and other clinical or laboratory research purposes within neurosurgery. Further studies comparing cost, speed, and accuracy of MinION with current NGS solutions are required to establish its clinical value.
Financial support and sponsorship
This study was supported by funds from the Barrow Neurological Foundation and the Newsome Chair in Neurosurgery Research held by Dr. Preul.
Conflicts of interest
There are no conflicts of interest.
References
1. . Available from: https://nanoporetech.com/community.
2. Burrows-Wheeler Aligner. Available from: (http://bio-bwa.sourceforge.net/.
3. . Available from: https://www.genscript.com/sequencing.html?src=pullmenu.
4. . Available from: https://usegalaxy.org/.
5. . Available from: https://metrichor.com.
6. . Available from: https://nanoporetech.com.
7. . Available from: https://bioconductor.org/packages/release/bioc/html/QDNAseq.html.
8. . Available from: https://cran.r-project.org.
9. Aibaidula A, Zhao W, Wu JS, Chen H, Shi ZF, Zheng LL. Microfluidics for rapid detection of isocitrate dehydrogenase 1 mutation for intraoperative application. J Neurosurg. 2016. 124: 1611-8
10. Ammar R, Paton TA, Torti D, Shlien A, Bader GD. Long read nanopore sequencing for detection of HLA and CYP2D6 variants and haplotypes. F1000Research. 2015. 4: 17-
11. Ashton PM, Nair S, Dallman T, Rubino S, Rabsch W, Mwaigwisya S. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat Biotechnol. 2015. 33: 296-300
12. Bi WL, Abedalthagafi M, Horowitz P, Agarwalla PK, Mei Y, Aizer AA. Genomic landscape of intracranial meningiomas. J Neurosurg. 2016. 125: 525-35
13. Chin CS, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A. Phased diploid genome assembly with single-molecule real-time sequencing. NatMethods. 2016. 13: 1050-4
14. Church G, Deamer DW, Branton D, Baldarelli R, Kasianowicz J. Google Patents, assignee. Characterization of individual polymer molecules based on monomer-interface interactions patent 5795782. 1998. p.
15. Cornelis S, Gansemans Y, Deleye L, Deforce D, Van Nieuwerburgh F. Forensic SNP Genotyping using Nanopore MinION Sequencing. Sci Rep. 2017. 7: 41759-
16. Cowperthwaite MC, Mohanty D, Burnett MG. Genome-wide association studies: Apowerful tool for neurogenomics. NeurosurgFocus. 2010. 28: E2-
17. David M, Dursi LJ, Yao D, Boutros PC, Simpson JT. Nanocall: An open source basecaller for Oxford Nanopore sequencing data. Bioinformatics (Oxford, England). 2017. 33: 49-55
18. De Mattos-Arruda L, Mayor R, Ng CK, Weigelt B, Martinez-Ricarte F, Torrejon D. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015. 6: 8839-
19. Delev D, Pavlova A, Grote A, Bostrom A, Hollig A, Schramm J. NOTCH4 gene polymorphisms as potential risk factors for brain arteriovenous malformation development and hemorrhagic presentation. J Neurosurg. 2017. 126: 1552-9
20. Euskirchen P, Bielle F, Labreche K, Kloosterman WP, Rosenberg S, Daniau M. Same-day genomic and epigenomic diagnosis of brain tumors using real-time nanopore sequencing. Acta Neuropathol. 2017. 134: 691-703
21. Ghali MG, Srinivasan VM, Mohan AC, Jones JY, Kan PT, Lam S. Pediatric cerebral cavernous malformations: Genetics, pathogenesis, and management. Surg Neurol Int. 2016. 7: S1127-34
22. Giampietro PF, Pourquie O, Raggio C, Ikegawa S, Turnpenny PD, Gray R. Summary of the first inaugural joint meeting of the International Consortium for scoliosis genetics and the International Consortium for vertebral anomalies and scoliosis, March 16-18, 2017, Dallas, Texas. Am J Med Genet A. 2018. 176: 253-6
23. Greninger AL, Naccache SN, Federman S, Yu G, Mbala P, Bres V. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 2015. 7: 99-
24. Havik AL, Bruland O, Myrseth E, Miletic H, Aarhus M, Knappskog PM. Genetic landscape of sporadic vestibular schwannoma. J Neurosurg. 2018. 128: 911-22
25. Hendrix P, Foreman PM, Harrigan MR, Fisher WS, Vyas NA, Lipsky RH. Endothelial Nitric Oxide Synthase Polymorphism Is Associated with Delayed Cerebral Ischemia Following Aneurysmal Subarachnoid Hemorrhage. World Neurosurg. 2017. 101: 514-9
26. Hong EP, Kim BJ, Kim C, Choi HJ, Jeon JP. Association of SOX17 Gene Polymorphisms and Intracranial Aneurysm: A Case-Control Study and Meta-Analysis. World Neurosurg. 2018. 110: e823-9
27. Hu J, Luo J, Chen Q. The Susceptibility Pathogenesis of Moyamoya Disease. World Neurosurg. 2017. 101: 731-41
28. Jain M, Koren S, Miga KH, Quick J, Rand AC, Sasani TA. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol. 2018. 36: 338-45
29. Jain M, Olsen HE, Paten B, Akeson M. The Oxford Nanopore MinION: Delivery of nanopore sequencing to the genomics community. Genome Biol. 2016. 17: 239-
30. Jansen HJ, Liem M, Jong-Raadsen SA, Dufour S, Weltzien FA, Swinkels W. Rapid de novo assembly of the European eel genome from nanopore sequencing reads. SciRep. 2017. 7: 7213-
31. Judge K, Hunt M, Reuter S, Tracey A, Quail MA, Parkhill J. Comparison of bacterial genome assembly software for MinION data and their applicability to medical microbiology. MicrobGenomics. 2016. 2: e000085-
32. Kilianski A, Haas JL, Corriveau EJ, Liem AT, Willis KL, Kadavy DR. Bacterial and viral identification and differentiation by amplicon sequencing on the MinION nanopore sequencer. Gigascience. 2015. 4: 12-
33. Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, Ganapathy G. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol. 2012. 30: 693-700
34. Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017. 27: 722-36
35. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J. Initial sequencing and analysis of the human genome. Nature. 2001. 409: 860-921
36. Laver T, Harrison J, O’Neill PA, Moore K, Farbos A, Paszkiewicz K. Assessing the performance of the Oxford Nanopore Technologies MinION. Biomol Detect Quantif. 2015. 3: 1-8
37. Li H. Minimap and miniasm: Fast mapping and de novo assembly for noisy long sequences. Bioinformatics (Oxford, England). 2016. 32: 2103-10
38. Li Z, Tan H, Shi Y, Huang G, Wang Z, Liu L. Global Gene Expression Patterns and Somatic Mutations in Sporadic Intracranial Aneurysms. World Neurosurg. 2017. 100: 15-21
39. Liu L, Li Y, Li S, Hu N, He Y, Pong R. Comparison of next-generation sequencing systems. J Biomed Biotechnol 2012. 2012. 251364:
40. Loman N. Available from: http://lab.loman.net/2017/03/09/ultrareads-for-nanopore/.
41. Loman NJ, Quick J, Simpson JT. A complete bacterial genome assembled de novo using only nanopore sequencing data. NatMethods. 2015. 12: 733-5
42. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016. 131: 803-20
43. Lu H, Giordano F, Ning Z. Oxford Nanopore MinION Sequencing and Genome Assembly. Genomics Proteomics Bioinformatics. 2016. 14: 265-79
44. Martirosyan NL, Patel AA, Carotenuto A, Kalani MY, Belykh E, Walker CT. Genetic Alterations in Intervertebral Disc Disease. Front Surg. 2016. 3: 59-
45. Minervini CF, Cumbo C, Orsini P, Anelli L, Zagaria A, Impera L. Mutational analysis in BCR-ABL1 positive leukemia by deep sequencing based on nanopore MinION technology. Exp MolPathol. 2017. 103: 33-7
46. Minervini CF, Cumbo C, Orsini P, Brunetti C, Anelli L, Zagaria A. TP53 gene mutation analysis in chronic lymphocytic leukemia by nanopore MinION sequencing. Diagn Pathol. 2016. 11: 96-
47. Mitsuhashi S, Kryukov K, Nakagawa S, Takeuchi JS, Shiraishi Y, Asano K. A portable system for rapid bacterial composition analysis using a nanopore-based sequencer and laptop computer. Sci Rep. 2017. 7: 5657-
48. Nakagawa Y, Sasaki H, Ohara K, Ezaki T, Toda M, Ohira T. Clinical and Molecular Prognostic Factors for Long-Term Survival of Patients with Glioblastomas in Single-Institutional Consecutive Cohort. World Neurosurg. 2017. 106: 165-73
49. Norris AL, Workman RE, Fan Y, Eshleman JR, Timp W. Nanopore sequencing detects structural variants in cancer. Cancer BiolTher. 2016. 17: 246-53
50. Pentsova EI, Shah RH, Tang J, Boire A, You D, Briggs S. Evaluating Cancer of the Central Nervous System Through Next-Generation Sequencing of Cerebrospinal Fluid. JClin Oncol. 2016. 34: 2404-15
51. Pfaff E, Kessler T, Balasubramanian GP, Berberich A, Schrimpf D, Wick A. Feasibility of real-time molecular profiling for patients with newly diagnosed glioblastoma without MGMT promoter-hypermethylation-the NCT Neuro Master Match (N2M2) pilot study. NeuroOncol. 2018. 20: 826-37
52. Quick J, Grubaugh ND, Pullan ST, Claro IM, Smith AD, Gangavarapu K. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc. 2017. 12: 1261-76
53. Quick J, Loman NJ, Duraffour S, Simpson JT, Severi E, Cowley L. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016. 530: 228-32
54. Quick J, Quinlan AR, Loman NJ. A reference bacterial genome dataset generated on the MinION portable single-molecule nanopore sequencer. Gigascience. 2014. 3: 22-
55. Rand AC, Jain M, Eizenga JM, Musselman-Brown A, Olsen HE, Akeson M. Mapping DNA methylation with high-throughput nanopore sequencing. Nat Methods. 2017. 14: 411-3
56. Sahm F, Schrimpf D, Jones DT, Meyer J, Kratz A, Reuss D. Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol. 2016. 131: 903-10
57. Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977. 74: 5463-7
58. Schatz MC, Delcher AL, Salzberg SL. Assembly of large genomes using second-generation sequencing. Genome Res. 2010. 20: 1165-73
59. Scheinin I, Sie D, Bengtsson H, van de Wiel MA, Olshen AB, van Thuijl HF. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res. 2014. 24: 2022-32
60. Shankar GM, Francis JM, Rinne ML, Ramkissoon SH, Huang FW, Venteicher AS. Rapid Intraoperative Molecular Characterization of Glioma. JAMA Oncol. 2015. 1: 662-7
61. Shendure J, Balasubramanian S, Church GM, Gilbert W, Rogers J, Schloss JA. DNA sequencing at 40: Past, present and future. Nature. 2017. 550: 345-53
62. Simon-Sanchez J, Singleton A. Genome-wide association studies in neurological disorders. Lancet Neurol. 2008. 7: 1067-72
63. Simpson JT, Workman RE, Zuzarte PC, David M, Dursi LJ, Timp W. Detecting DNA cytosine methylation using nanopore sequencing. NatMethods. 2017. 14: 407-10
64. Sovic I, Sikic M, Wilm A, Fenlon SN, Chen S, Nagarajan N. Fast and sensitive mapping of nanopore sequencing reads with GraphMap. NatCommun. 2016. 7: 11307-
65. Suzuki A, Suzuki M, Mizushima-Sugano J, Frith MC, Makalowski W, Kohno T. Sequencing and phasing cancer mutations in lung cancers using a long-read portable sequencer. DNA Res. 2017. 24: 585-96
66. Teng H, Hall MB, Duarte T, Cao MD, Coin L. Chiron: Translating nanopore raw signal directly into nucleotide sequence using deep learning. Gigascience. 2018. 7:
67. Tyson JR, O’Neil NJ, Jain M, Olsen HE, Hieter P, Snutch TP. MinION-based long-read sequencing and assembly extends the Caenorhabditis elegans reference genome. Genome Res. 2018. 28: 266-74
68. Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG. The sequence of the human genome. Science (New York, NY). 2001. 291: 1304-51
69. Wang Y, Springer S, Zhang M, McMahon KW, Kinde I, Dobbyn L. Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc Natl Acad Sci U S A. 2015. 112: 9704-9
70. Wick RJudd LMHolt KELast accessed on 2018 Jun 24. Available from: https://github.com/rrwick/Basecallingcomparison/tree/v3.0.
71. Worst BC, van Tilburg CM, Balasubramanian GP, Fiesel P, Witt R, Freitag A. Next-generation personalised medicine for high-risk paediatric cancer patients-The INFORM pilot study. EurJ Cancer (Oxford, England: 1990. 2016. 65: 91-101
72. Wu Y, Li Z, Shi Y, Chen L, Tan H, Wang Z. Exome Sequencing Identifies LOXL2 Mutation as a Cause of Familial Intracranial Aneurysm. World Neurosurg. 2018. 109: e812-8
73. Xu Z, Li H, Song J, Han B, Wang Z, Cao Y. Meta-Analysis of Microarray-Based Expression Profiles to Identify Differentially Expressed Genes in Intracranial Aneurysms. World Neurosurg. 2017. 97: 661-8 e667