- Department of Neurological Surgery, University of Washington, Seattle, Washington, United States.
- Genomic Medicine Group, J. Craig Venter Institute, La Jolla, California, United States.
Correspondence Address:
Melanie Walker, Department of Neurological Surgery, University of Washington, Seattle, Washington, United States.
DOI:10.25259/SNI_104_2022
Copyright: © 2022 Surgical Neurology International This is an open-access article distributed under the terms of the Creative Commons Attribution-Non Commercial-Share Alike 4.0 License, which allows others to remix, transform, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.How to cite this article: Melanie Walker1, Carla Uranga2, Samuel HS Levy1, Cory Kelly1, Anna Edlund2. Thrombus-associated microbiota in acute ischemic stroke patients. 10-Jun-2022;13:247
How to cite this URL: Melanie Walker1, Carla Uranga2, Samuel HS Levy1, Cory Kelly1, Anna Edlund2. Thrombus-associated microbiota in acute ischemic stroke patients. 10-Jun-2022;13:247. Available from: https://surgicalneurologyint.com/surgicalint-articles/11643/
Abstract
Background: Despite a reduction in stroke incidence and age-standardized death rates, stroke remains a leading cause of death and disability worldwide. Significant interest in recent years has focused on the microbiota-host interaction because accumulating evidence has revealed myriad ways in which bacteria may contribute to risk of stroke and adverse outcomes after stroke. The emergence of endovascular thrombectomy as a treatment provides a unique opportunity to utilize thrombus retrieved from cerebral arteries to fill knowledge gaps about the influence of bacteria on stroke pathophysiology. While bacterial signatures have been confirmed in cerebral thrombi, the exact nature of the pathogenesis has not been established.
Methods: Thrombi were obtained from a cohort of adult ischemic stroke patients during standard of care thrombectomy. After DNA extraction and quantification, thrombi underwent 16S rRNA amplicon-based metagenomic sequencing, followed by bioinformatics processing. Taxonomic identification of bacterial colonies isolated on Agar plates from plated suspension was performed using DNA extraction and full length 16S Sanger sequencing.
Results: A broad diversity of bacterial signatures was identified in specimens, primarily of cariogenic origin.
Conclusion: In this small study, we demonstrate proof of concept and technical feasibility for amplicon-based metagenomic sequencing of arterial thrombi and briefly discuss preliminary findings, challenges, and near-term translational opportunities for thrombus genomics.
Keywords: Metagenomics, Microbiota, Stroke, Thrombectomy
INTRODUCTION
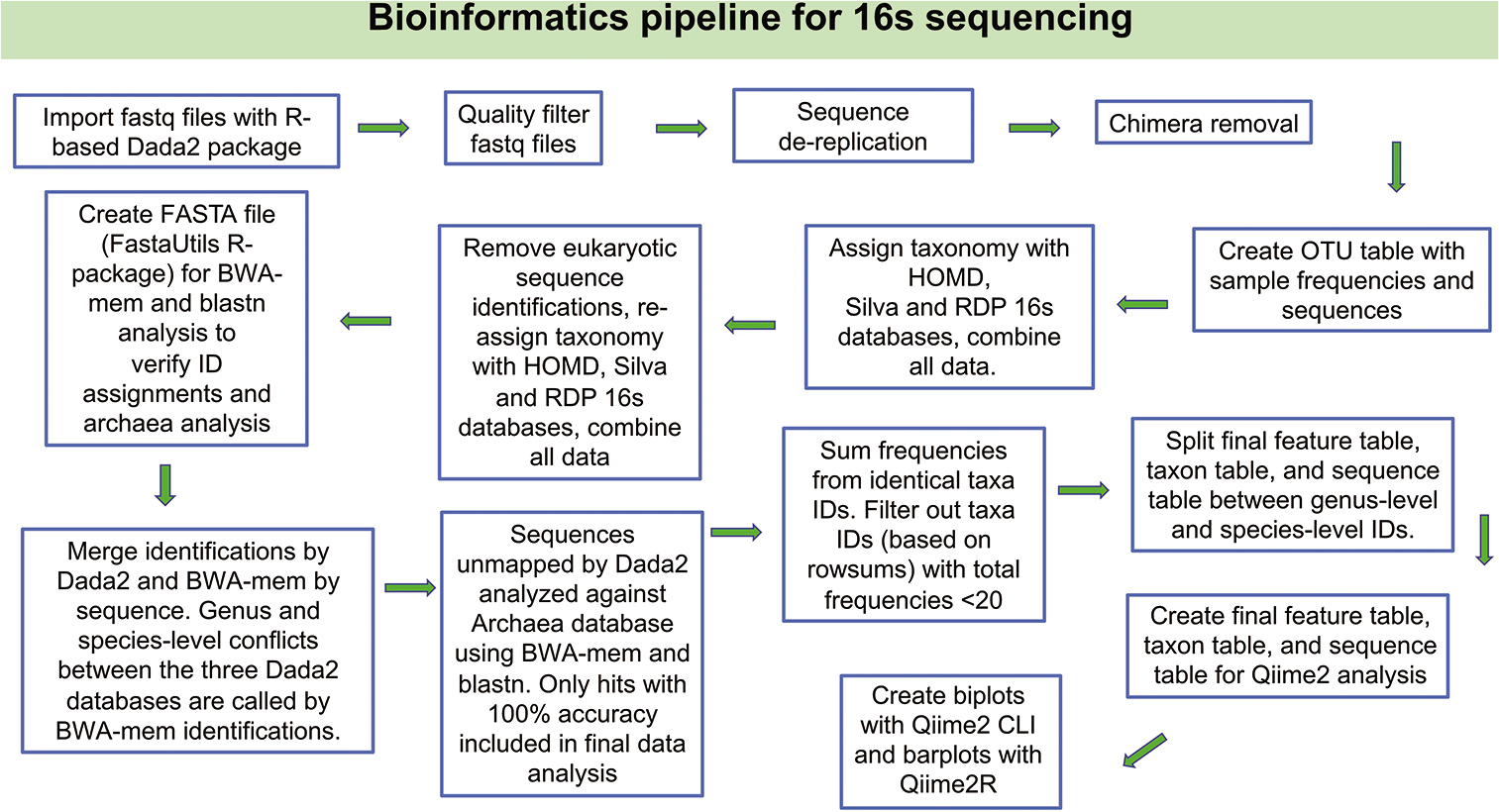
Accumulating evidence suggests that the human microbiota may influence the development or outcomes related to acute ischemic stroke (AIS). Cerebral thrombi represent a new source of biological information and may provide insight into the vascular microenvironment. No standards or guidelines exist for next generation sequencing (NGS) of thrombus-associated microbiota. To demonstrate proof of concept and technical feasibility in this setting, cerebral thrombi were subjected to amplicon-based bacterial 16S rRNA gene sequencing.
MATERIALS AND METHODS
Materials
Subjects, procedure, and specimens
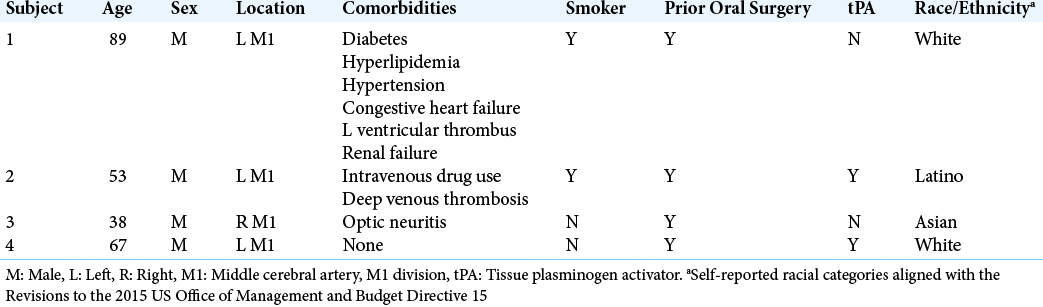
Thrombi were obtained from cerebral arteries of subjects over age 18 with AIS during standard-of-care endovascular thrombectomy (EVT) at a Comprehensive Stroke Center between January 2020 and October 2020. Analysis was limited to subjects whose thrombi were obtained intact, with full reperfusion (mTICI = 3).[
During the thrombectomy, a combined technique was used. In addition to stent retriever, aspiration was applied through an intermediate catheter as well as a large-bore guide catheter positioned in the cervical carotid to help ensure first pass reperfusion. Because the thrombus is frequently adherent to the devices, it must be washed with sterile saline and gently manipulated for transfer into the sterile collection receptacle. The thrombus was rinsed with sterile water, placed into a pre-prepared sterile collection container containing phosphate buffered saline (PBS), and stored at −80°C until processing.
Methods
DNA extraction and quantification
After homogenization, DNA extraction was performed using the Ultra-Deep Microbiome Prep protocol (Molzym, Portland, OR). DNA was quantified using Qubit® dsDNA high-sensitivity and broad-range fluorometric assays (Thermo Fisher Scientific, Waltham, MA). DNA quality was assessed using Agilent 2100 Bioanalyzer technology (Agilent, Santa Clara, CA).
Analysis of thrombus microbiota and quality control
Multiplex 16S rRNA gene (16S) fragment amplification (2 × 300bp paired end) using the Swift Amplicon® panel (SWIFT Biosciences, Ann Arbor, MI) that targets all nine variable regions (V1-V9) of bacterial and archaeal 16S was performed, followed by Swift 2S® Turbo DNA library preparation (SWIFT Biosciences) and deep sequencing through MiSeq® platform (Illumina Inc. La Jolla, CA). Negative controls (water) were included in all sample preparation and sequencing experiments. The R-based DADA2 open-source software package next-generation microbiome bioinformatics algorithm[
RESULTS
Subjects
The mean age of subjects was 62, with a standard deviation of 18.8 years. Although all were male, four unique racial/ ethnicities were represented.[
Bacterial DNA identified in cerebral thrombi
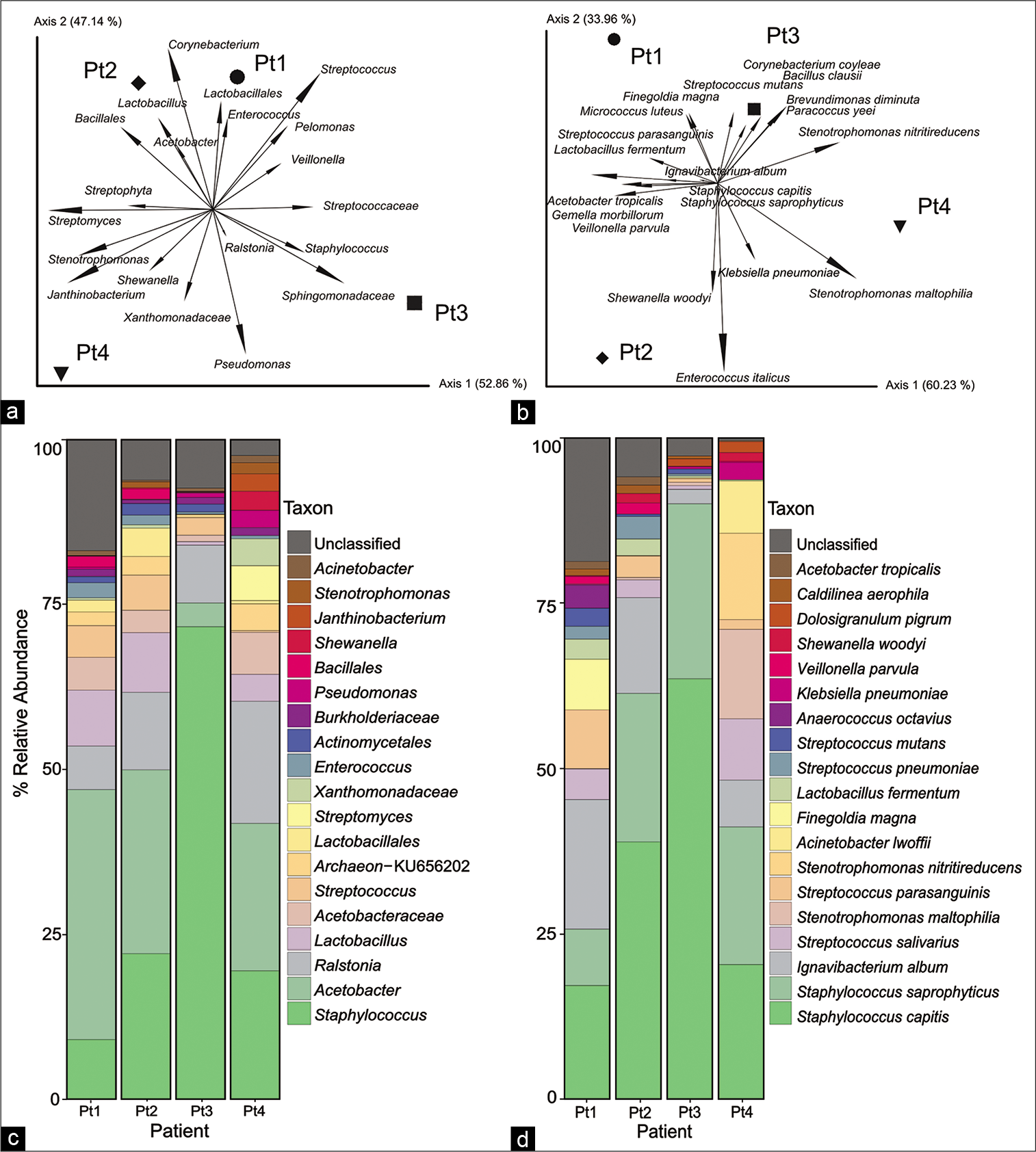
Deep sequencing of forward and reverse sequence reads representing all nine variable regions of the bacterial 16S amplified from DNA revealed the presence of bacterial signatures. The main bacterial groups associated with specimens in this cohort belonged to the Acetobacter, Streptococcus, and Lactobacillus genera [
Figure 2:
Biplot analysis of bacterial taxa based on 16S fragment sequencing (a and b) reveals the presence of a diverse bacterial community within cerebral thrombi. Signatures of oral pathogens, specifically cariogenic Staphylococcus, Streptococcus, and Lactobacillus were revealed in thrombi (c and d).
DISCUSSION
Validation of prior reports
Analysis of the thrombus-associated microbiota in this cohort supports findings from several previous studies, which report oral bacterial signatures in cerebral,[
Technical challenges
Several technical challenges were present, including low biomass specimens, insufficient specimen controls, and sample preservation conditions. Low-biomass samples with high non-microbial (host) nucleic acids may yield small quantities of bacterial DNA that may be insufficient for library construction.[
CONCLUSION
Analysis of the thrombus microbiota from cerebral thrombi is technically feasible and demonstrated the presence of multiple taxonomic groups. The confirmation of bacterial signatures representing the human microbiota, specifically the oral microbiota,[
Declaration of patient consent
Institutional Review Board (IRB) permission obtained for the study.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
1. Adams HP, Bendixen BH, Kappelle LJ, Biller J, Love BB, Gordon DL. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial TOAST. Trial of Org 10172 in acute stroke treatment. Stroke. 1993. 24: 35-41
2. Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019. 37: 852-7
3. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016. 13: 581-3
4. Caufield PW, Schön CN, Saraithong P, Li Y, Argimón S. Oral lactobacilli and dental caries: A model for niche adaptation in humans. J Dent Res. 2015. 94: 110S-8S
5. De Paoli P. Biobanking in microbiology: From sample collection to epidemiology, diagnosis and research. FEMS Microbiol Rev. 2005. 29: 897-910
6. Deo PN, Deshmukh R. Oral microbiome: Unveiling the fundamentals. J Oral Maxillofac Pathol. 2019. 23: 122-8
7. Farhangi MA, Vajdi M, Asghari-Jafarabadi M. Gut microbiota-associated metabolite trimethylamine N-Oxide and the risk of stroke: A systematic review and dose-response meta-analysis. Nutr J. 2020. 19: 76
8. Forssten SD, Björklund M, Ouwehand AC. Streptococcus mutans caries and simulation models. Nutrients. 2010. 2: 290-8
9. Gerber JC, Miaux YJ, von Kummer R. Scoring flow restoration in cerebral angiograms after endovascular revascularization in acute ischemic stroke patients. Neuroradiology. 2015. 57: 227-40
10. Guyard A, Boyez A, Pujals A, Robe C, Tran Van Nhieu J, Allory Y. DNA degrades during storage in formalin-fixed and paraffin-embedded tissue blocks. Virchows Arch. 2017. 471: 491-500
11. Kunin V, Copeland A, Lapidus A, Mavromatis K, Hugenholtz P. A bioinformatician’s guide to metagenomics. Microbiol Mol Biol Rev. 2008. 72: 557-78
12. Lee SB, Crouse CA, Kline MC. Optimizing storage and handling of DNA extracts. Forensic Sci Rev. 2010. 22: 131-44
13. Lyden P, Raman R, Liu L, Emr M, Warren M, Marler J. National institutes of health stroke scale certification is reliable across multiple venues. Stroke. 2009. 40: 2507-11
14. McCormack MG, Smith AJ, Akram AN, Jackson M, Robertson D, Edwards G. Staphylococcus aureus and the oral cavity: An overlooked source of carriage and infection?. Am J Infect Control. 2015. 43: 35-7
15. Office of Management and Budget (OMB) Standards. Available from: https://orwh.od.nih.gov/toolkit/other-relevant-federal-policies/OMB-standards [Last accessed on 30 Apr 20].
16. Patrakka O, Pienimäki JP, Tuomisto S, Ollikainen J, Lehtimäki T, Karhunen PJ. Oral bacterial signatures in cerebral thrombi of patients with acute ischemic stroke treated with thrombectomy. J Am Heart Assoc. 2019. 8: e012330
17. Pessi T, Karhunen V, Karjalainen PP, Ylitalo A, Airaksinen JK, Niemi M. Bacterial signatures in thrombus aspirates of patients with myocardial infarction. Circulation. 2013. 127: 1219-28
18. Rai SN, Qian C, Pan J, Rai JP, Song M, Bagaitkar J. Microbiome data analysis with applications to pre-clinical studies using QIIME2: Statistical considerations. Genes Dis. 2021. 8: 215-23
19. Reinhardt C. The gut microbiota as an influencing factor of arterial thrombosis. Hamostaseologie. 2019. 39: 173-9
20. Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014. 12: 87
21. Vajpeyee A, Chauhan PS, Pandey S, Tiwari S, Yadav LB, Shroti AK. Metagenomics analysis of thrombus samples retrieved from mechanical thrombectomy. Neurointervention. 2021. 16: 39-45
22. Vakhitov D, Tuomisto S, Martiskainen M, Korhonen J, Pessi T, Salenius JP. Bacterial signatures in thrombus aspirates of patients with lower limb arterial and venous thrombosis. J Vasc Surg. 2018. 67: 1902-7
23. Weyrich LS, Farrer AG, Eisenhofer R, Arriola LA, Young J, Selway CA. Laboratory contamination over time during low-biomass sample analysis. Mol Ecol Resour. 2019. 19: 982-96
24. Zaidat OO, Castonguay AC, Linfante I, Gupta R, Martin CO, Holloway WE. First pass effect: A new measure for stroke thrombectomy devices. Stroke. 2018. 49: 660-6
25. Zhou X, Nanayakkara S, Gao JL, Nguyen KA, Adler CJ. Storage media and not extraction method has the biggest impact on recovery of bacteria from the oral microbiome. Sci Rep. 2019. 9: 14968